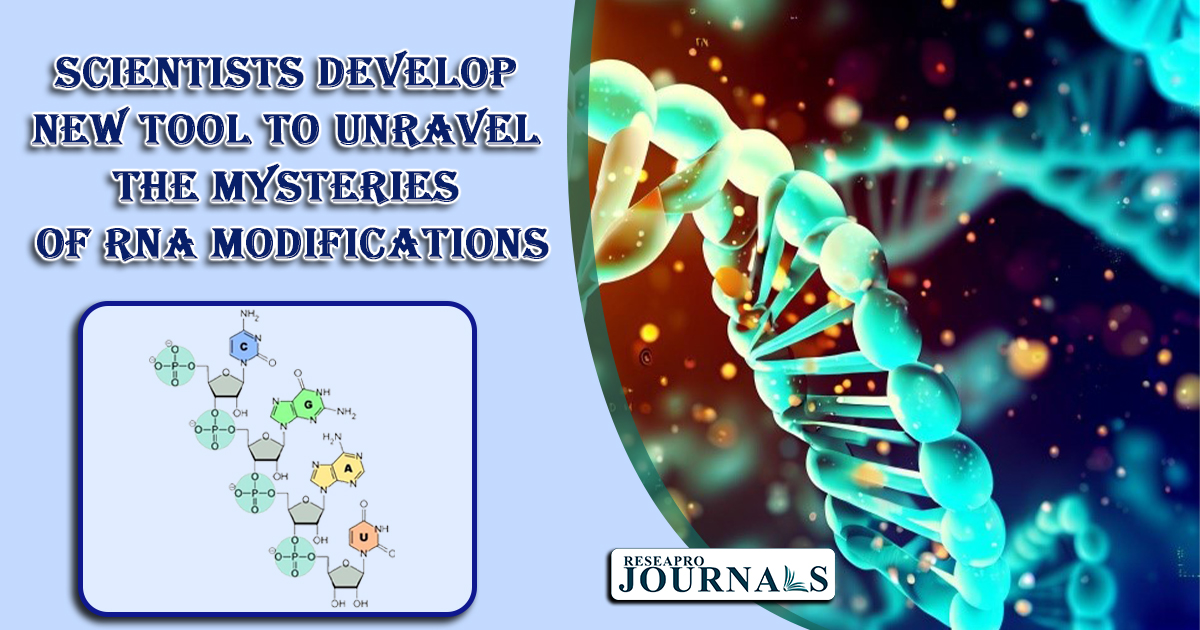
A new tool called iMVP has been developed for the visualization and interpretation of epitranscriptomic data. iMVP uses a nonlinear dimension reduction technique and density-based partition to subtype, visualize, and denoise epitranscriptomic signals.
iMVP has been shown to be effective in identifying previously unknown RNA modification motifs and writers, discovering false positives that are undetectable by traditional methods, and comparing different approaches to epitranscriptomic profiling.
iMVP could be a valuable tool for researchers who are studying the role of RNA modifications in disease, such as cancer. By helping scientists to better understand RNA modifications, iMVP could lead to the development of new diagnostic and therapeutic tools.